Mitochondrial Dynamics
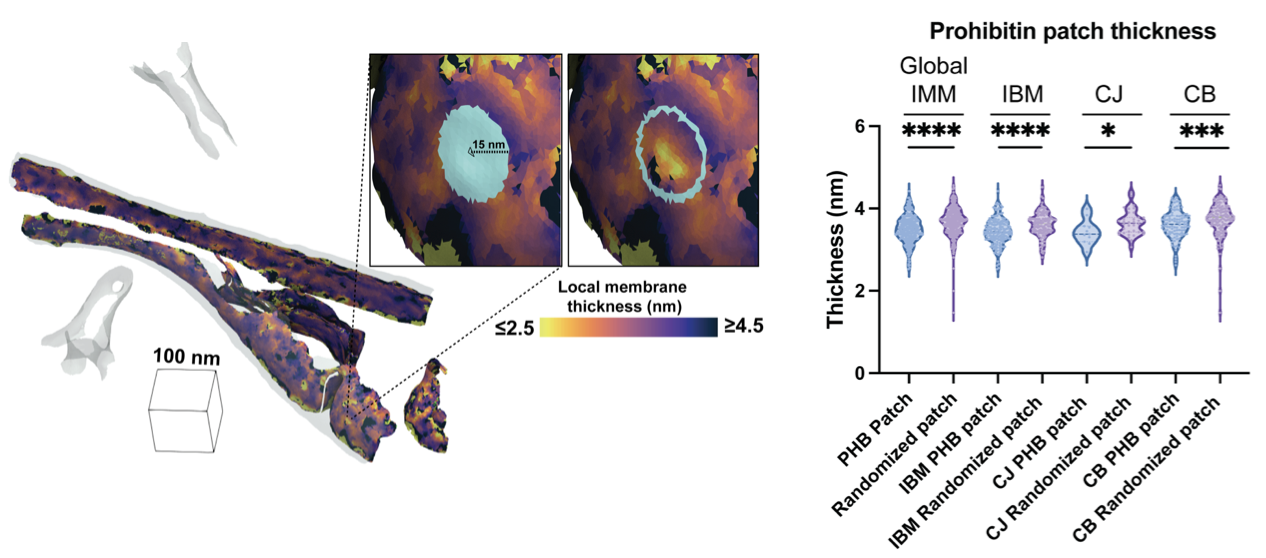
Medina M, Rahmani H, Chang Y, Barad BA, Grotjahn DA§. Prohibitin complexes associate with unique membrane microdomains in cells. bioRxiv 2025.
-
Access the paper
- PMID: XXXX
- Biorxiv Preprint: 688579
- Full Text Additional Links
- Surface Morphometrics on Github
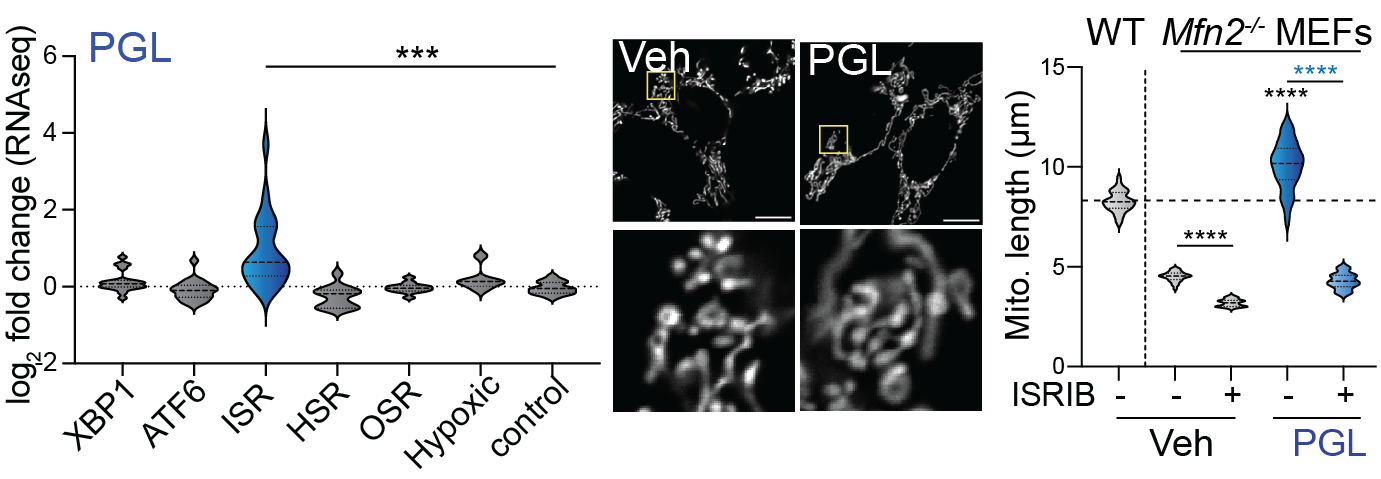
Bora P†, Zaman M†, Oviedo S, Kutseikin S, Madrazo N, Mathur P, Pannikkat M, Krasny S, Aldakhlallah R, Chu A, Johnson KA, Grotjahn DA, Shutt TE§, Wiseman RL§. Drug Repurposing Screen Identifies an HRI Activating Compound that Promotes Adaptive Mitochondrial Remodeling in MFN2-deficient Cells, PNAS 2025.†Co-first authors, §Co-corresponding authors.
-
Access the paper
- PMID: 41289394
- Biorxiv Preprint: 651574
- Full Text Additional Links
- Wiseman Lab
- Shutt Lab
Chang Y, Barad BA, Rahmani H, Zid BM, Grotjahn DA§. Cytoplasmic ribosomes on mitochondria alter the local membrane environment for protein import. Journal of Cell Biology (JCB) 2025.§Corresponding author
-
Access the paper
- PMID: 39071314
- Biorxiv Preprint: 604013
- Full Text
- Deposited Maps: 48751, 48752 Additional Links
- Selected article for The Year in Cell Biology: 2025
- Press Release
- Zid Lab
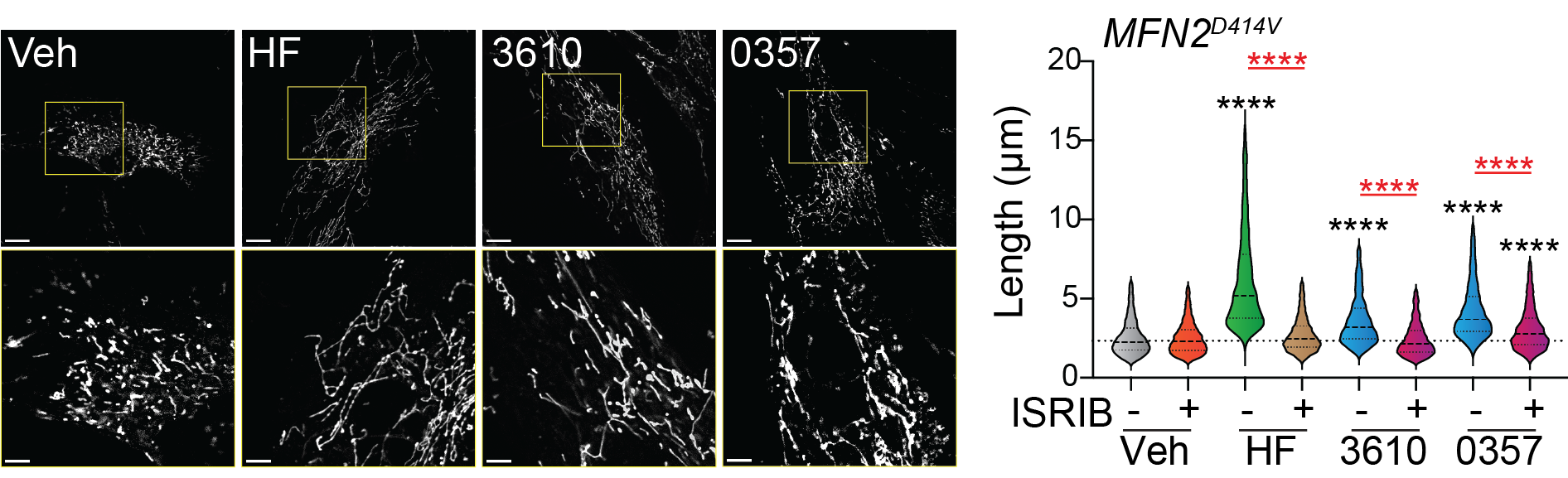
Baron KR†, Oviedo S† Krasny S, Zaman M, Aldakhlallah R, Mathur P, Pfeffer G, Bollong MJ, Shutt T, Grotjahn DA§ and Wiseman RL§. Pharmacologic activation of integrated stress response kinases inhibits pathologic mitochondrial fragmentation. eLife 2024 †Equal contribution §Corresponding author
-
Access the paper
- PMID: 38915623
- Biorxiv Preprint: 598126
- Full Text Additional Links
- Wiseman Lab @ Scripps Research
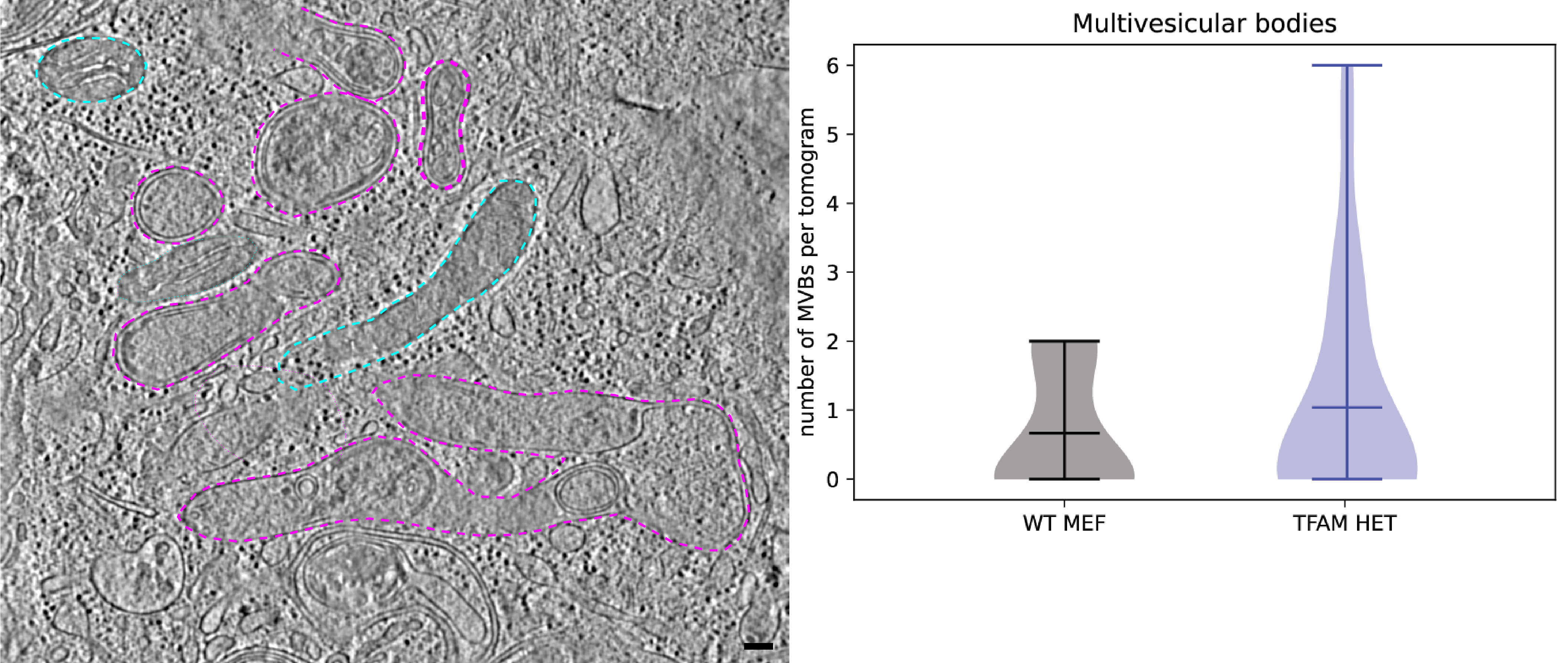
Newman LE, Tadepalle N, Weiser Novak S, Rojas GR, Tadepalle N, Schiavon CR, Grotjahn DA, Towers CG, Tremblay M, Donnelly MP, Ghosh S, Medina M, Rocha S, Rodriguez-Enriquez R, Chevez JA, Lemersal I, Manor U, Shadel GS. Mitochondrial DNA replication stress triggers a pro-inflammatory endosomal pathway of nucleoid disposal. Nature Cell Biology 2024
-
Access the paper
- PMID: 38332353
- Biorxiv Preprint: 511955
- Full Text Additional Links
- Shadel Lab
- Manor Lab
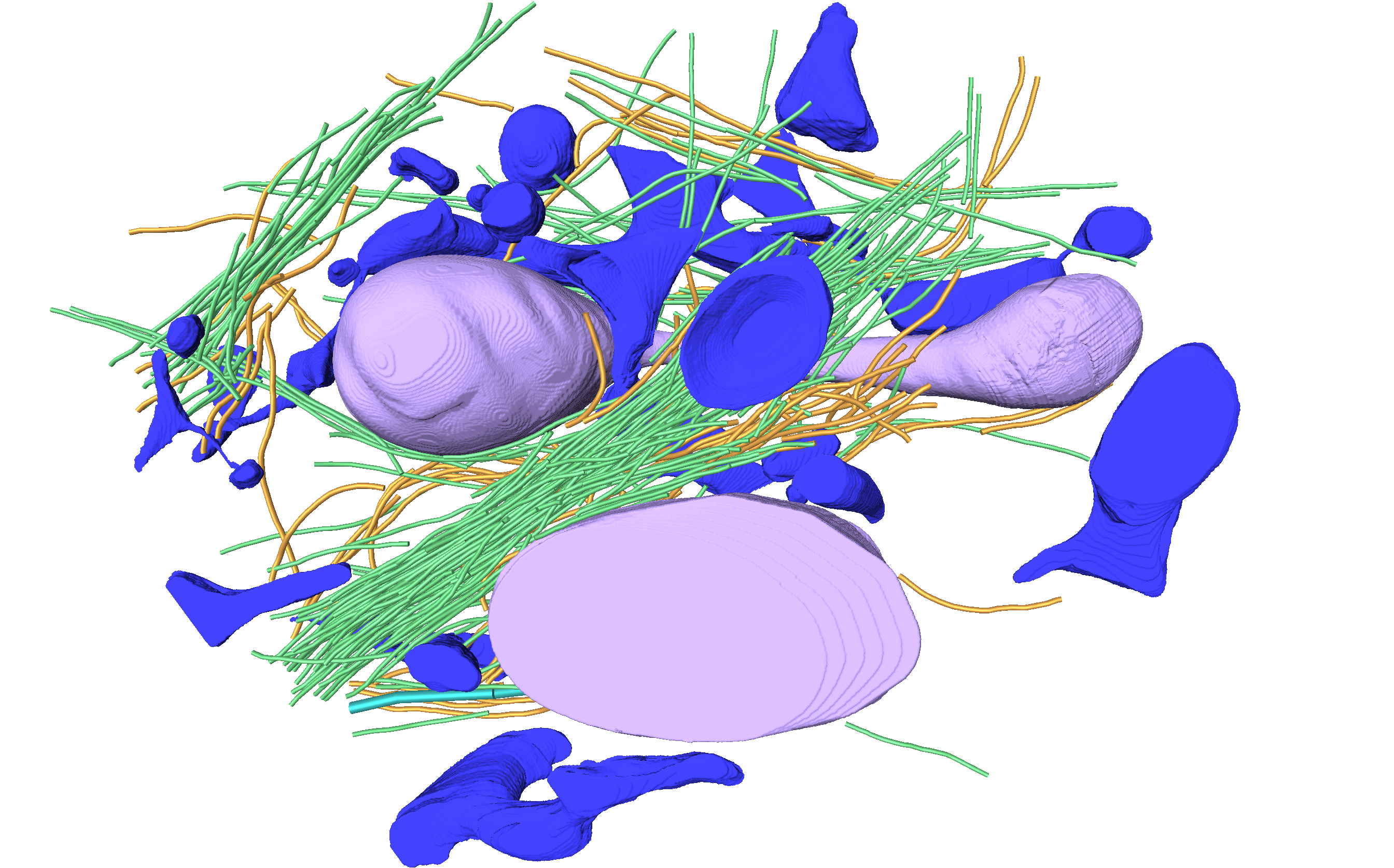
Mageswaran SK†, Grotjahn DA†§, Zeng X, Barad BA, Medina M, Hoang MH, Dobro MJ, Chang YW, Xu M, Yang WY, Jensen, GJ§ . Nanoscale details of mitochondrial constriction revealed by cryo-electron tomography. Biophysical Journal 2023 †Equal contribution §Corresponding authors
-
Access the paper
- PMID: 37533259
- Biorxiv Preprint: 472487
- Full Text Additional Links
- scripts for quantifying filament architectures
- Jensen Lab
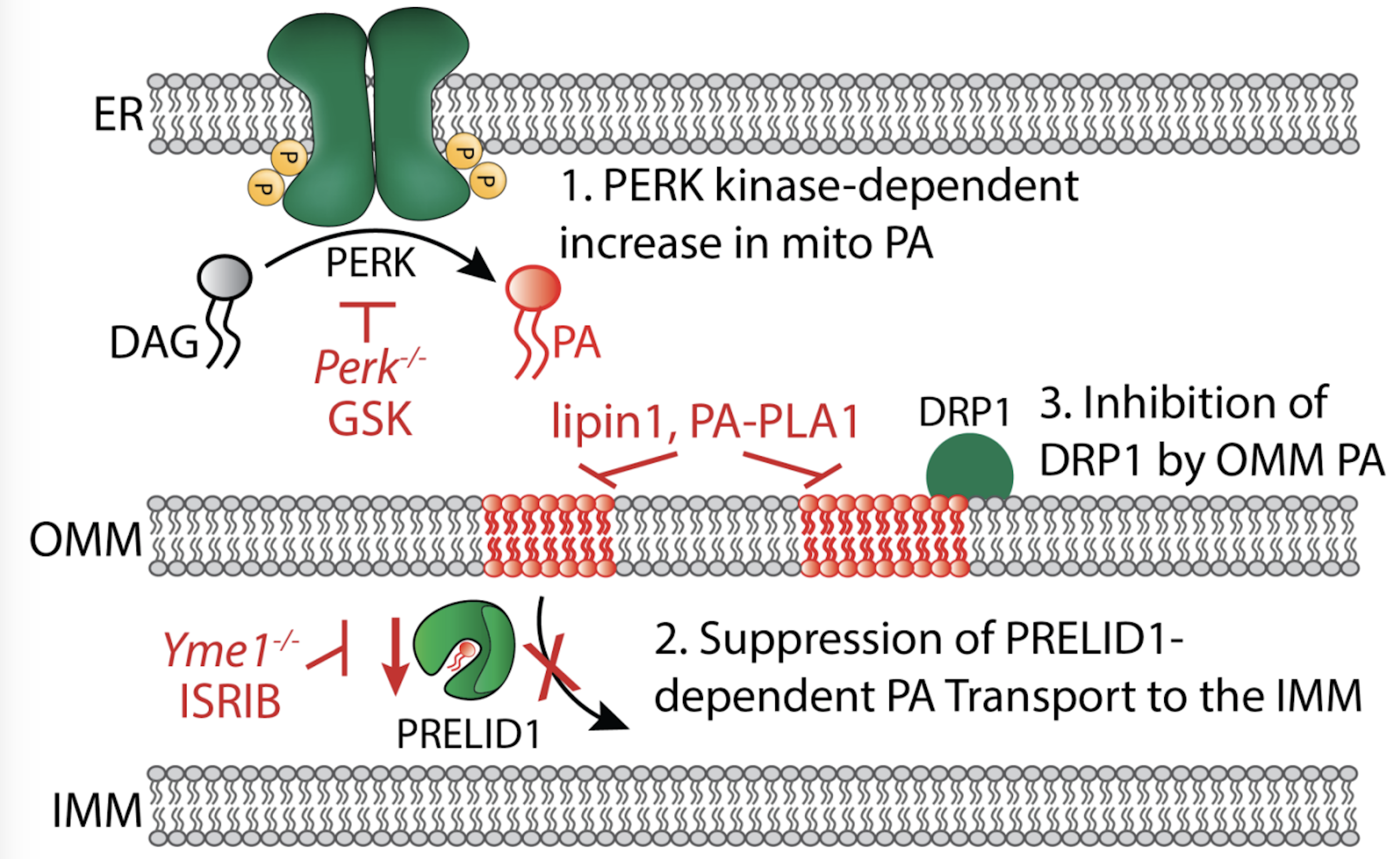
Perea V, Cole C, Lebeau J, Dolina V, Baron KR, Madhavan A, Kelly JW, Grotjahn DG, Wiseman RL. PERK signaling promotes mitochondrial elongation by remodeling membrane phosphatidic acid. The EMBO Journal 2023
-
Access the paper
- PMID: 37306086
- Biorxiv Preprint: 481593
- Full Text Additional Links
- Wiseman Lab
Surface Morphometrics and other cryo-ET software
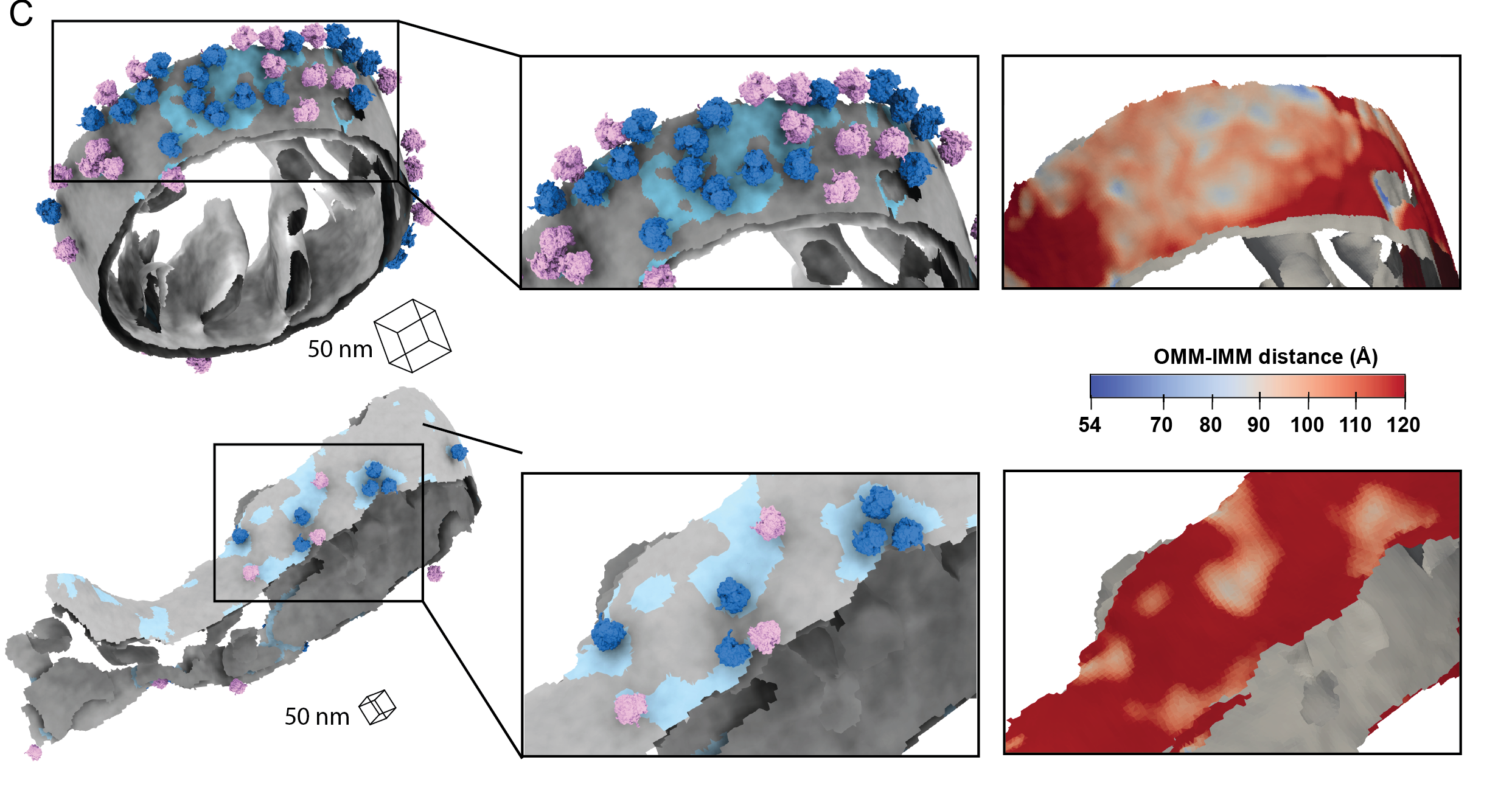
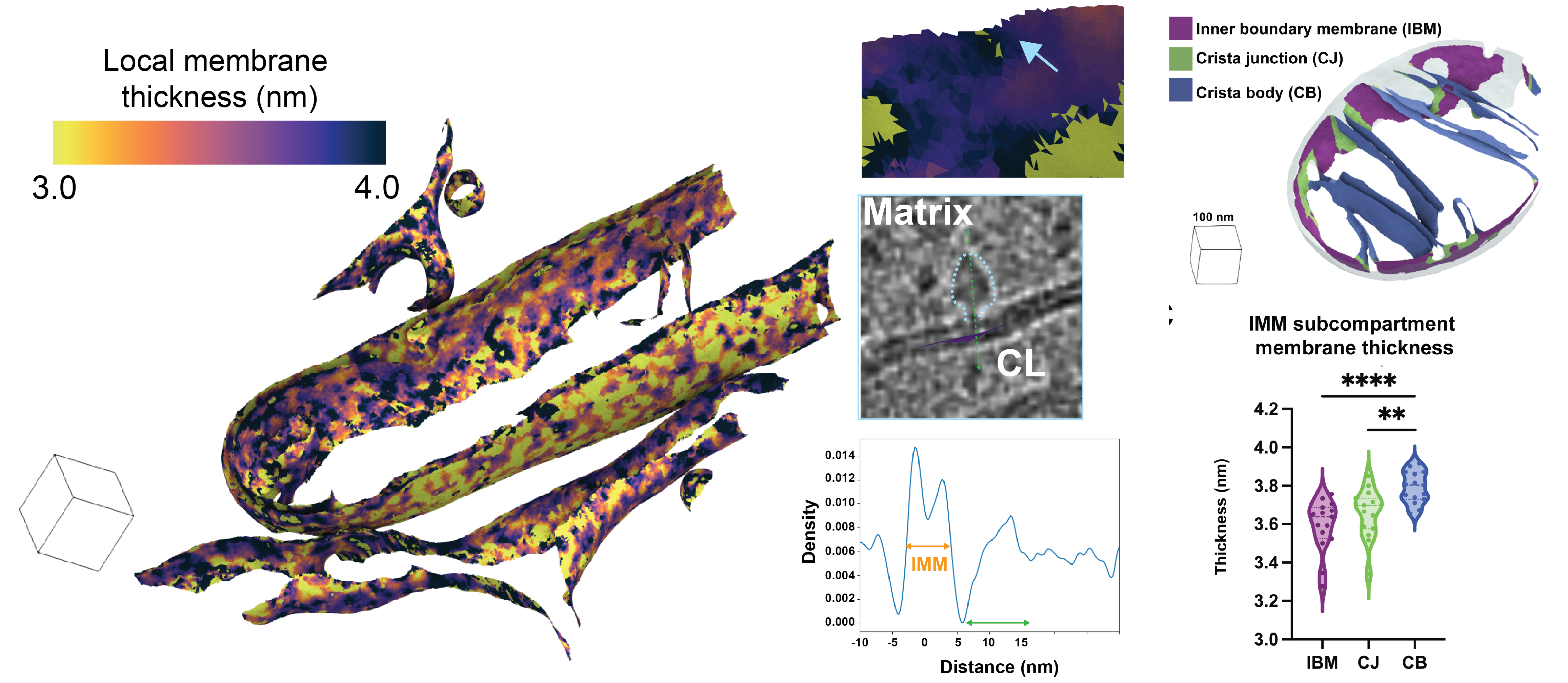
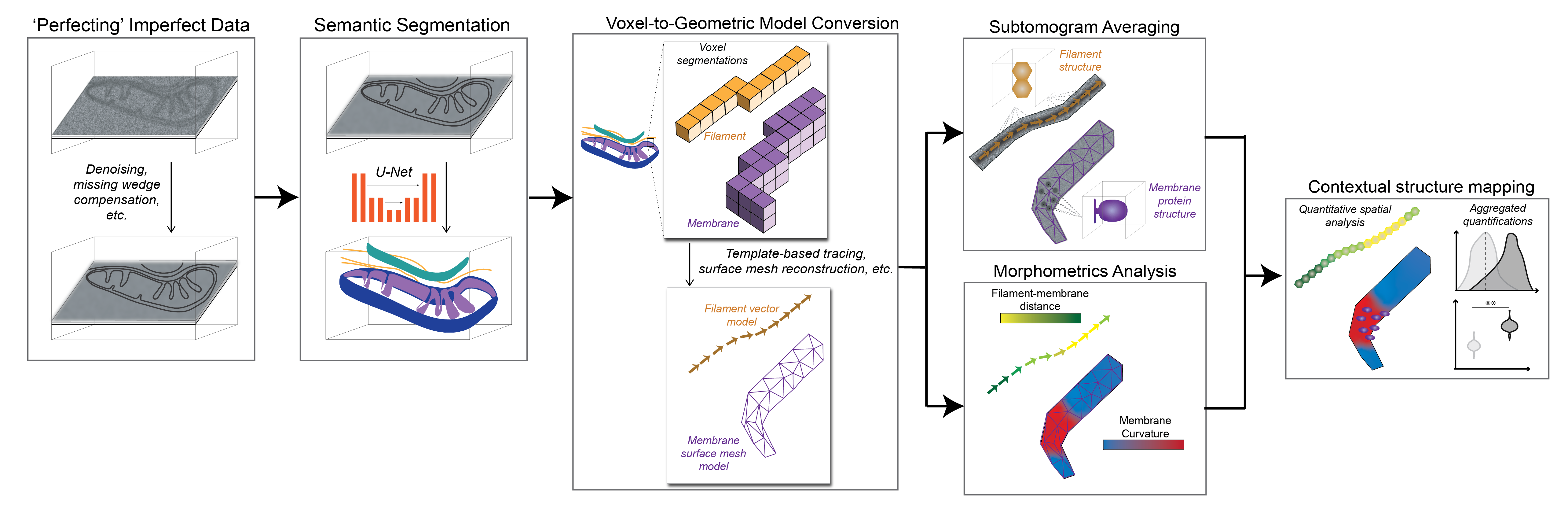
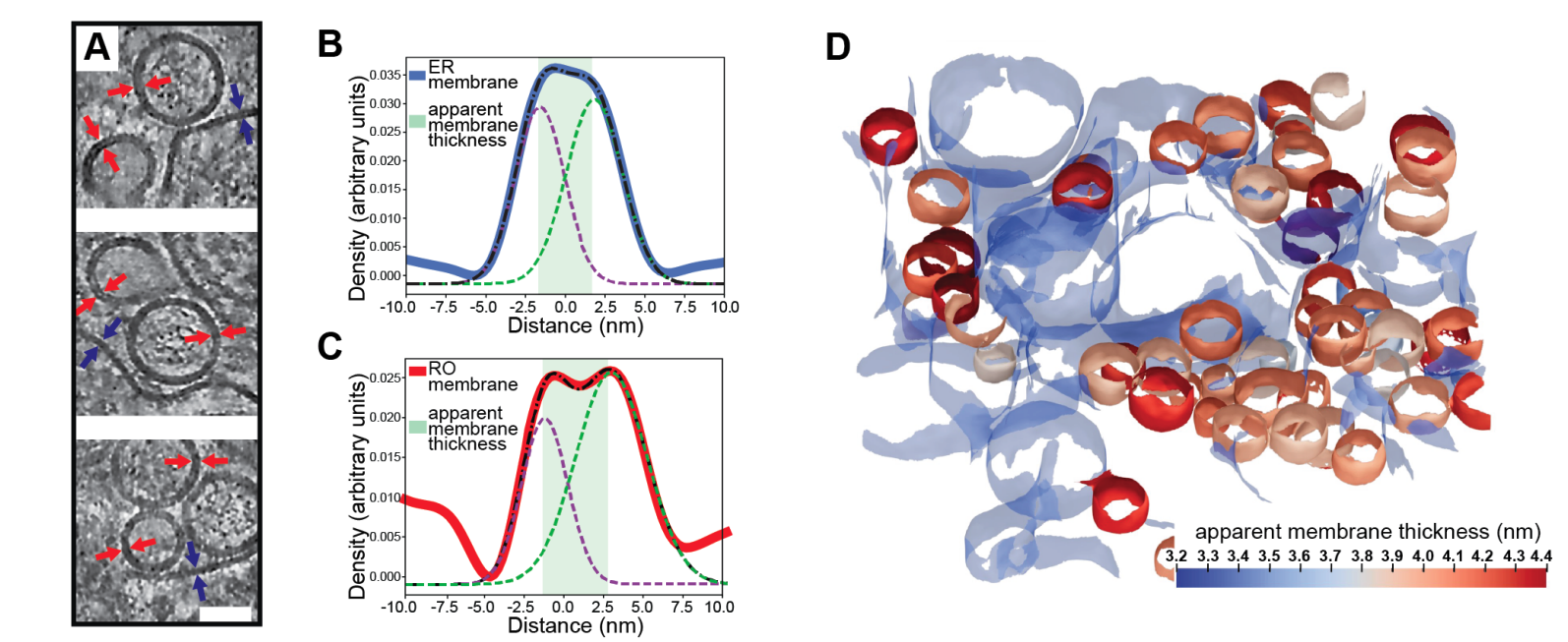
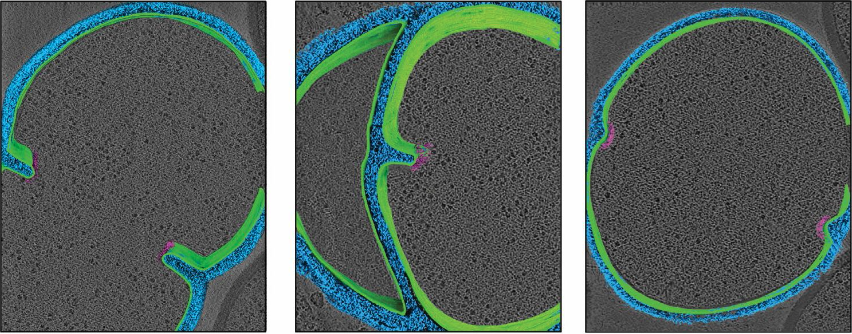
Medina M†, Chang Y†, Rahmani H, Frank M, Khan Z, Fuentes D, Heberle FA, Waxham MN, Barad BA§, Grotjahn DA§. Surface Morphometrics reveals local membrane thickness variation in organellar subcompartments. Journal of Cell Biology (JCB) 2025.†Equal contribution §Corresponding author
-
Access the paper
- PMID: 40654968
- Biorxiv Preprint: 651574
- Full Text
- Deposited Map: 72321 Additional Links
- Surface Morphometrics on Github
- Patch analysis on Github
- EMPIAR Deposition (Tilt Series, Tomograms, Segmentations, and Surfaces)
- Barad Lab
Grotjahn DA. Segmenting cryo-electron tomography data: Extracting models from cellular landscapes. Current Opinion in Structural Biology 2025.
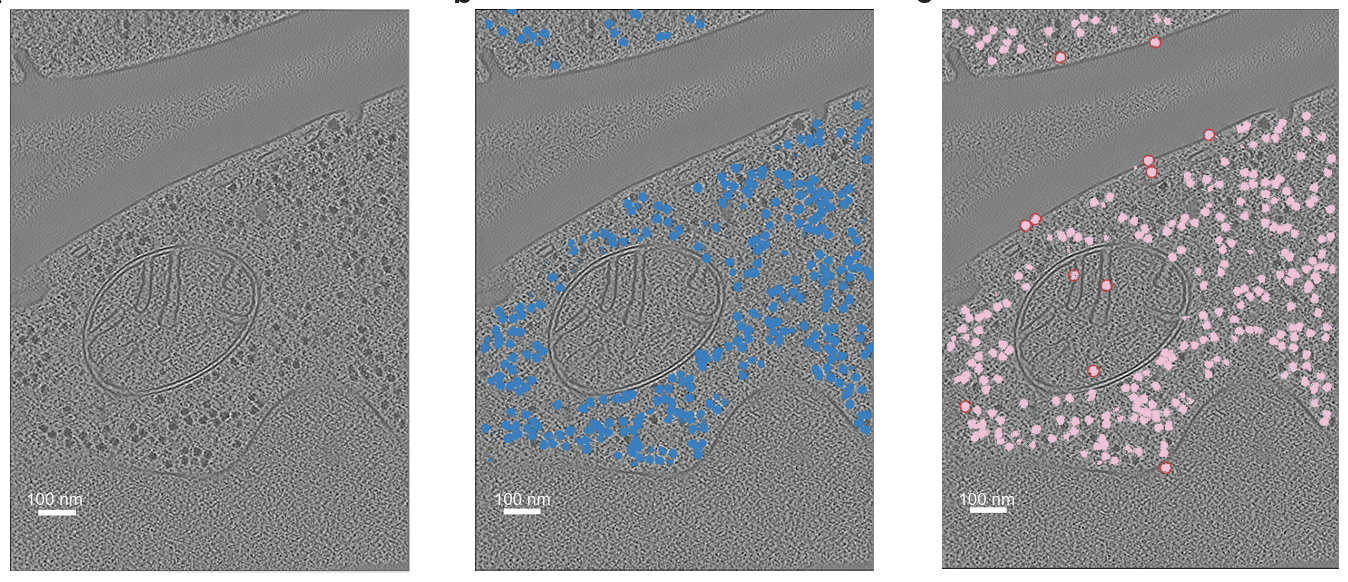
Hosseini R, Liang Y, Singh D, Rahmani H, Choe D, Lee J, Xu M, Segal E, Zou J, Williamson J, Grotjahn DA, Villa E, Xie P. RobPicker: A Meta Learning Framework for Robust Identification of Macromolecules in Cryo-Electron Tomograms. bioRxiv 2025.
-
Access the paper
- PMID: XXXX
- Biorxiv Preprint: 676650
- Full Text Additional Links
- Pengtao Xie Lab
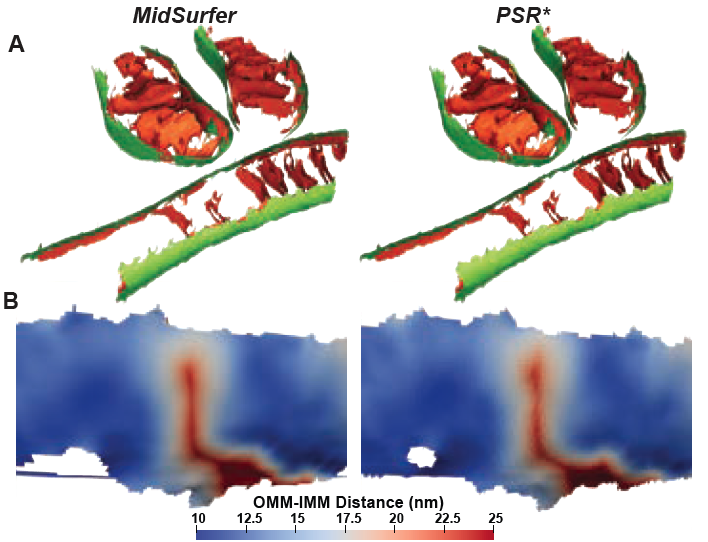
Boneš E, Khan D, Bohak C, Barad BA, Grotjahn DA, Viola I, Theußl T. MidSurfer: A parameter-free approach for mid-surface extraction from segmented volumetric data. IEEE 2025
-
Access the paper
- PMID: 41129462
- arXiv preprint: 2405.19339
- Full Text Additional Links
- MidSurfer on Github
- KAUST Visualization Core Lab
- KAUST NANOVIS: Nanovisualization Research Group
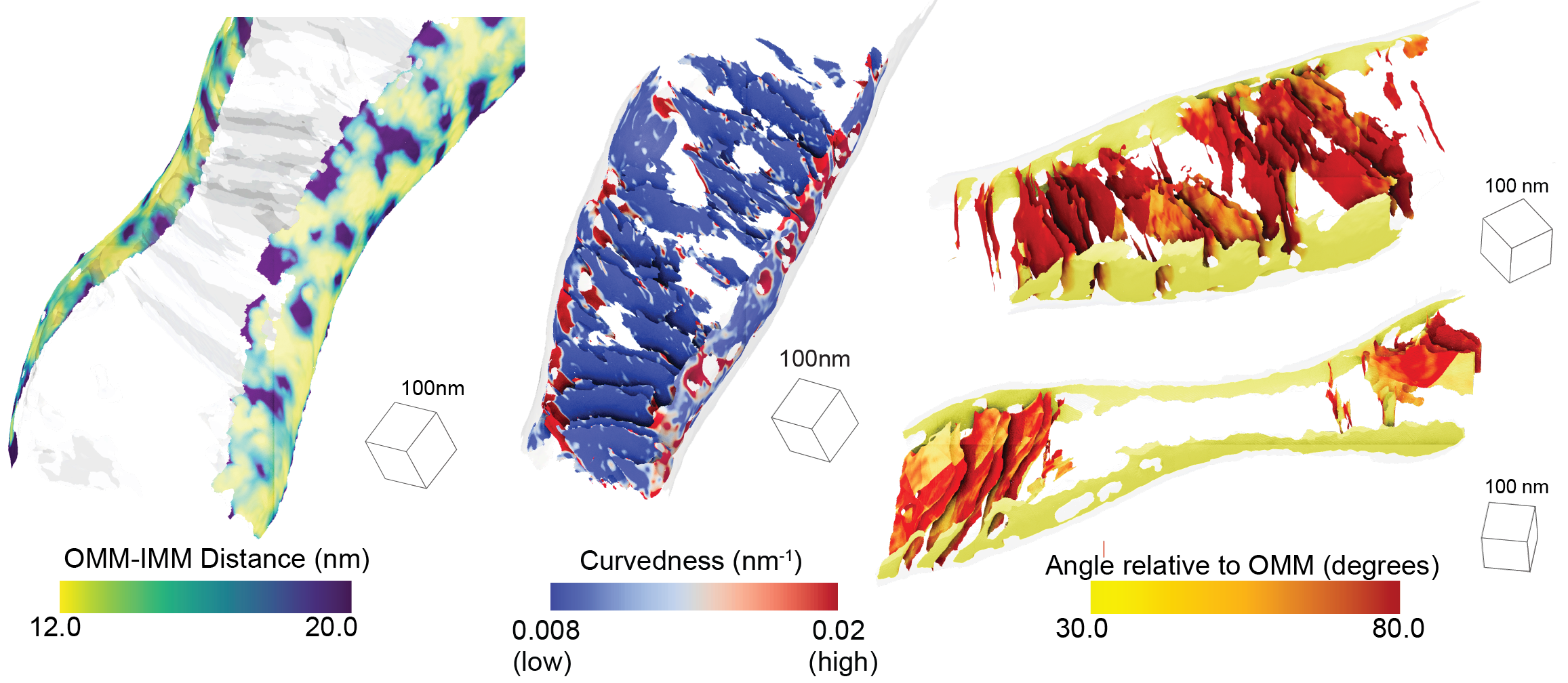
Barad BA†, Medina M†, Fuentes D, Wiseman, RL, Grotjahn DA§. Quantifying organellar ultrastructure in cryo-electron tomography using a surface morphometrics pipeline. Journal of Cell Biology (JCB) 2023 †Equal contribution §Corresponding author
-
Access the paper
- PMID: 36786771
- Biorxiv Preprint: 477440
- Full Text Additional Links
- Press Release
- Journal Tweetorial
- Preprint Tweetorial
- Surface Morphometrics on Github
- EMPIAR Deposition (Tilt Series, Tomograms, Segmentations, and Surfaces)
- Zenodo Deposition (Quantifications and Statistics)
- Review Commons Reviews
- Wiseman Lab @ Scripps Research
Gardner A, Ludovic A, Fuentes D, Maritan M, Barad BA, Medina, M, Olson AJ, Grotjahn DA, Goodsell DS. CellPAINT: Turnkey illustration of molecular cell biology. Frontiers in Bioinformatics 2021
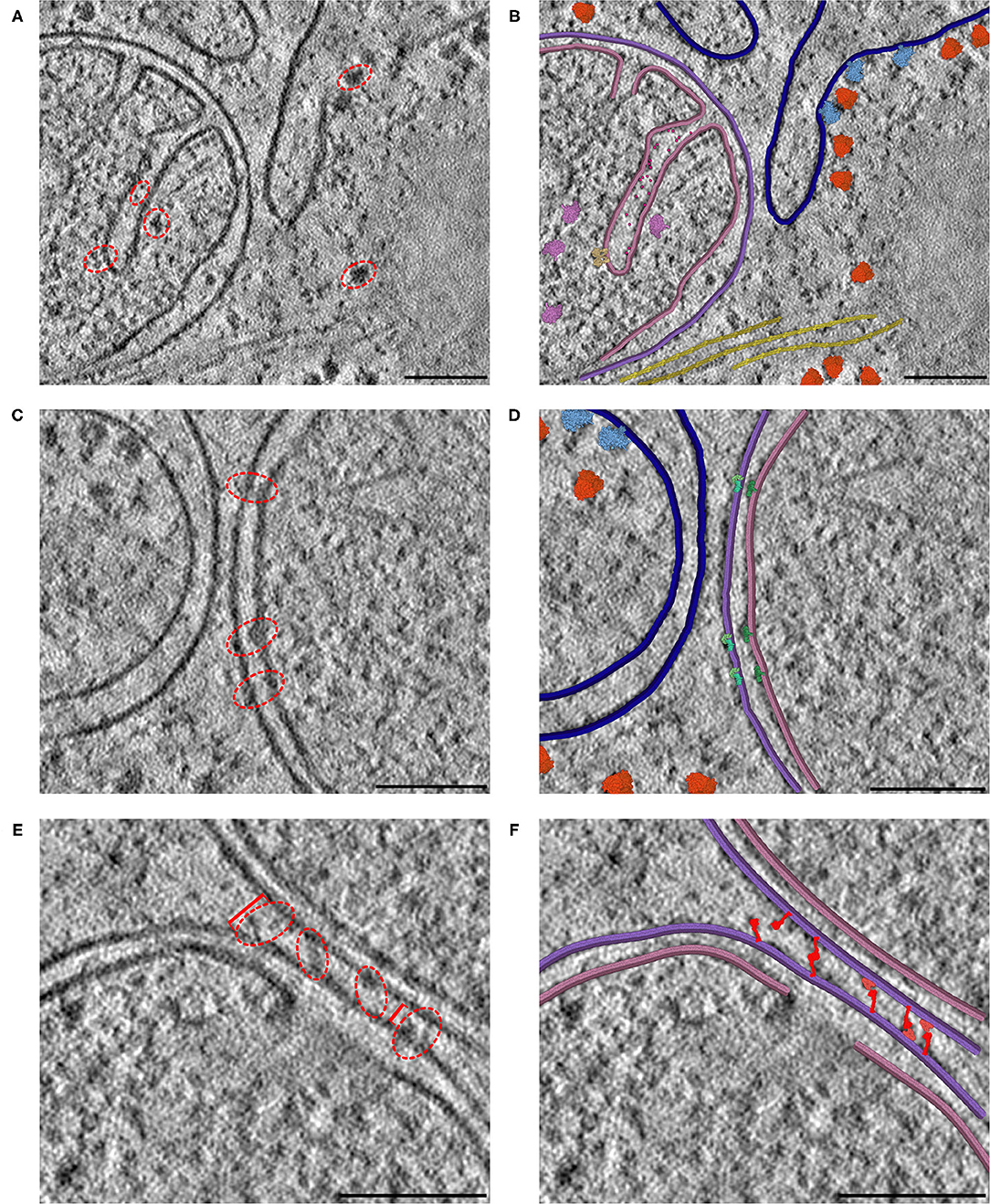
-
Access the paper
- PMID: 34790910
- PMCID: PMC8594902
- Full Text
- Zenodo Record:
(Tomogram used for CellPAINT hypothesis generation)
Additional Links
- Molecular Graphics Laboratory
Basanta B, Chowdhury S, Lander GC§, Grotjahn DA§. A guided approach for subtomogram averaging of challenging macromolecular assemblies. Journal of Structural Biology: X 2020 §Corresponding authors
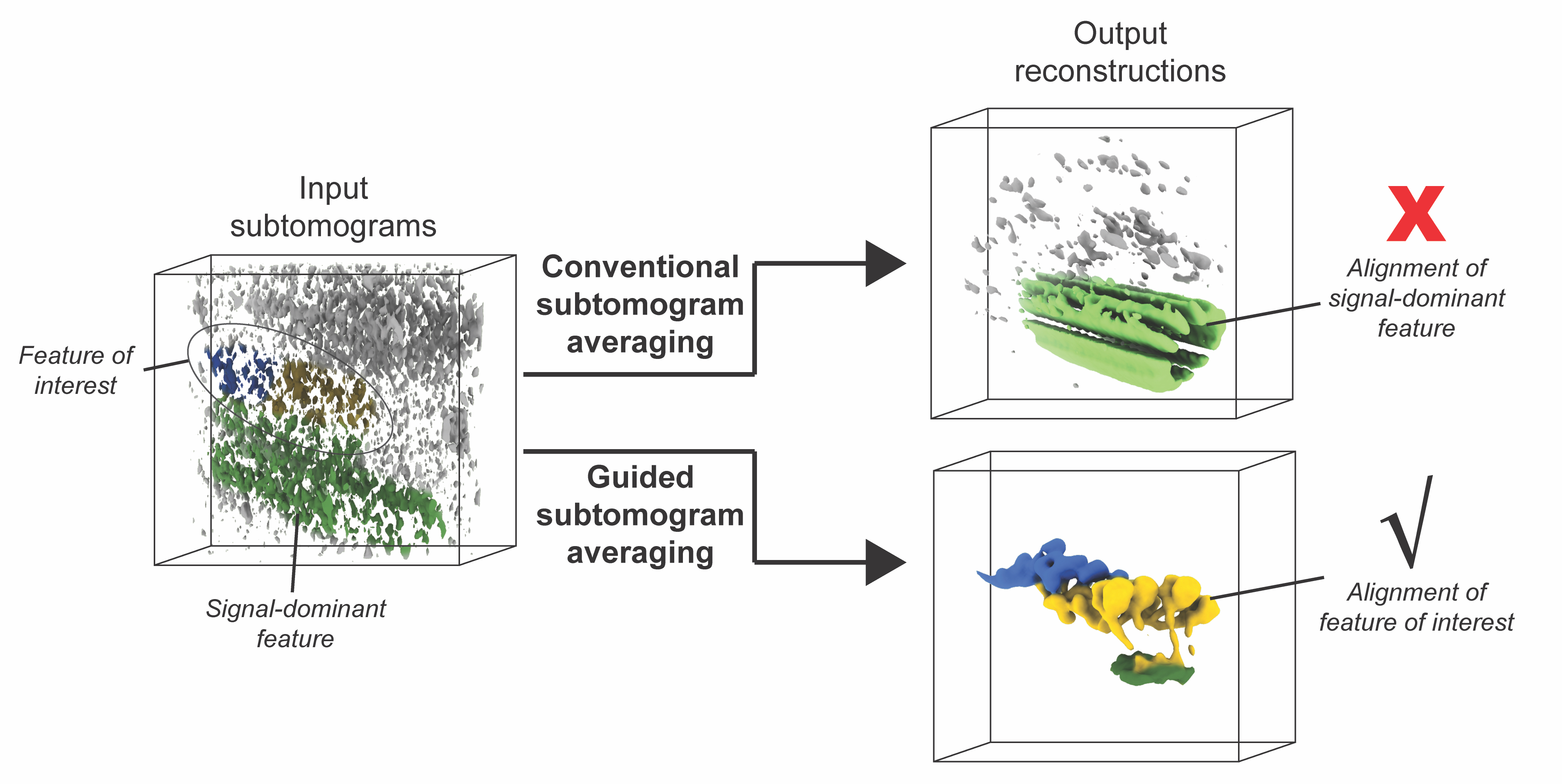
-
Access the paper
- PMID: 33319208
- PMCID: PMC7724198
- Biorxiv Preprint: 930297
- Full Text Additional Links
- guided_tomo_align scripts
- Lander Lab
Cytoskeletal motors and filaments
Mahalingan KK, Grotjahn DA, Li Y, Lander GC, Zehr EA, and Roll-Mecak A. Structural basis for alpha-tubulin-specific and modification history dependent glutamate chain elongation by tubulin tyrosine ligase-like family enzymes. Nature Chemical Biology 2024
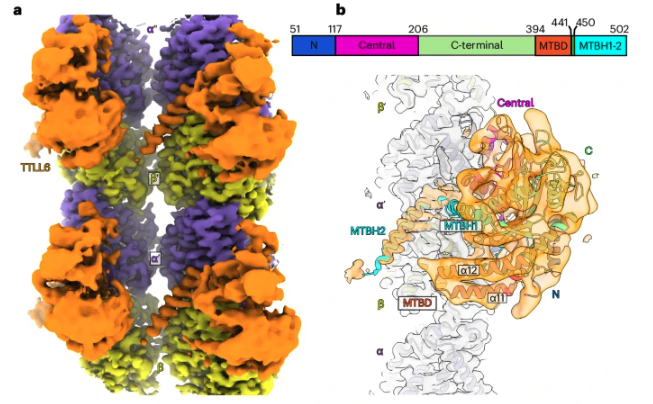
-
Access the paper
- PMID: 38658656
- Full Text Additional Links
- Roll-Mecak Lab
- Lander Lab
Grotjahn DA and Lander GC. Setting the dynein motor in motion: New insights from electron tomography. Journal of Biological Chemistry (JBC) 2019.
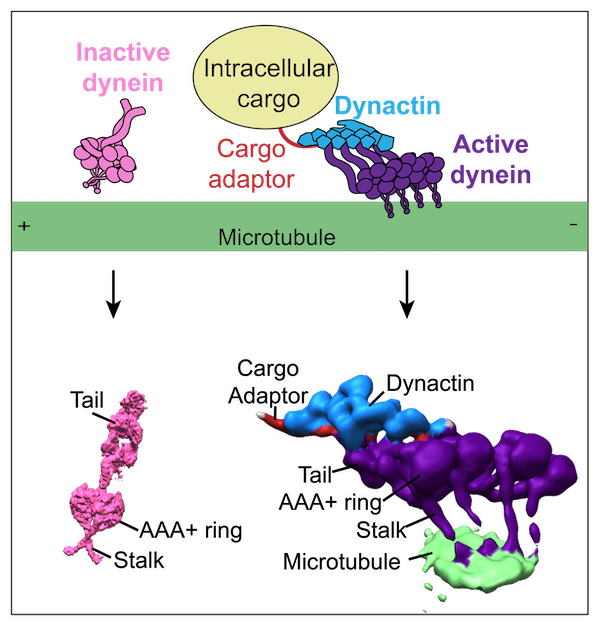
-
Access the paper
- PMID: 31285262
- PMCID: PMC6737236
- Full Text Additional Links
- Lander Lab
Grotjahn DA, Chowdhury S, Xu Y, McKenney RJ, Schroer TA, Lander GC. Cryo-electron tomography reveals that dynactin recruits a team of dyneins for processive motility. Nature Structural and Molecular Biology 2018.
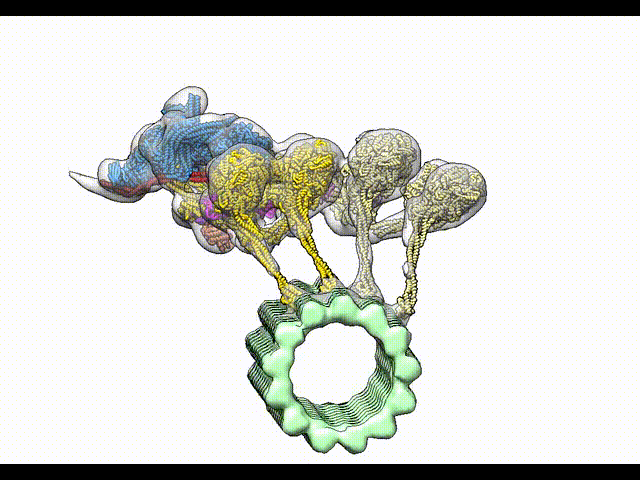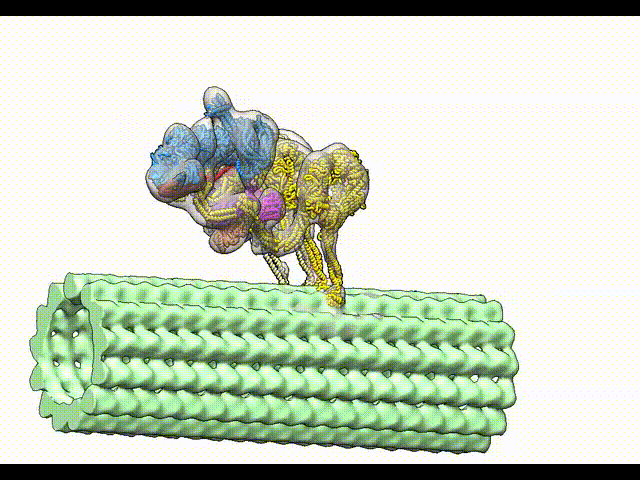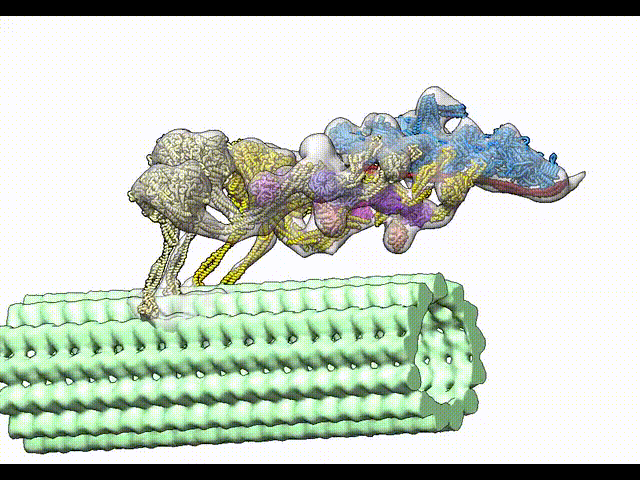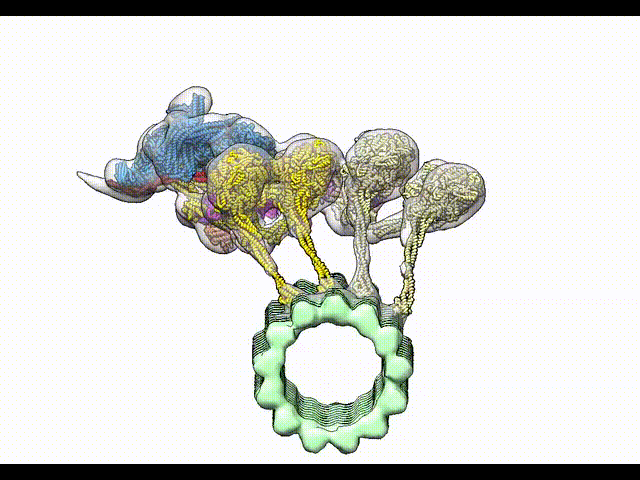
-
Access the paper
- PMID: 29416113
- PMCID: PMC5969528
- Biorxiv Preprint: 182576
- Full Text
- Deposited Maps: 7000, 7001 Additional Links
- Lander Lab
- F1000Prime
Ge X, Grotjahn DA, Welch E, Lyman-Gingerich J, Holguin C, Dimitrova E, Abrams EW, Gupta T, Marlow FL, Yabe T, Adler A, Mullins MC, Pelegri F. Hecate/Grip2a acts to reorganize the cytoskeleton in the symmetry-breaking event of embryonic axis induction. PLOS Genetics 2014.
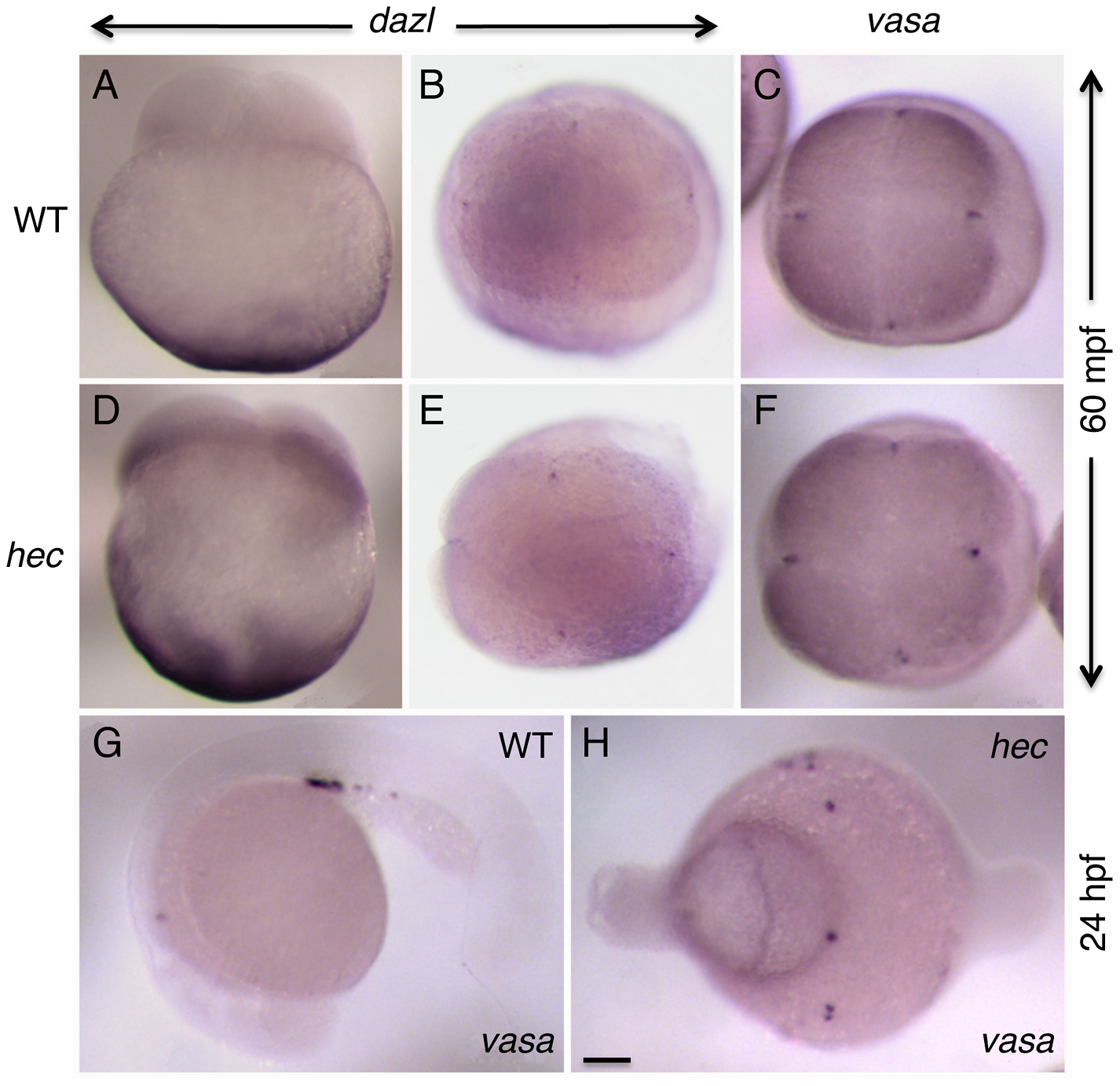
-
Access the paper
- PMID: 24967891
- PMCID: PMC4072529
- Full Text
Cryo-ET across biological systems
Rodriguez ZK, JAndino-Moncada JR, Buth SA, Mehrani A, Ranaweera A, Shi J, Andrade LR, Singh SP, Strutzenberg TS, Marin M, Guerrero-Ferreira R, Rahmani H, Grotjahn DA, Stagg S, Melikyan GB, Lyumkis D, Francis AC. Time-Resolved Fluorescence Imaging and Correlative Cryo-Electron Tomography to Study Structural Changes of the HIV-1 Capsid. ACS Nano 2025.
-
Access the paper
- PMID: 40844114
- Full Text Additional Links
- Lyumkis Lab
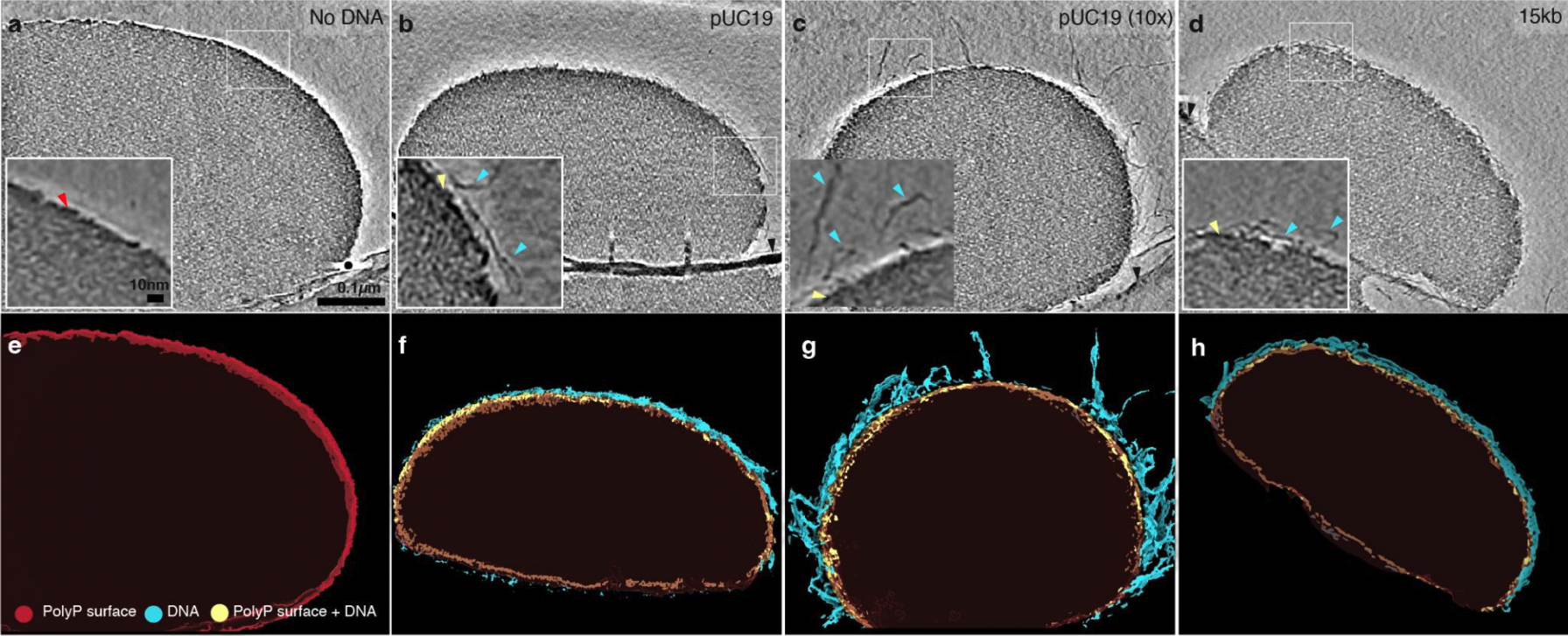
Chawla R†, Tom JKA†, Boyd T, Tu NH, Bai T Grotjahn DA, Park D, Deniz AA, Racki LR. Reentrant DNA shellts tune polyphosphate condensate size. Nature Communications 2024 †Equal contribution
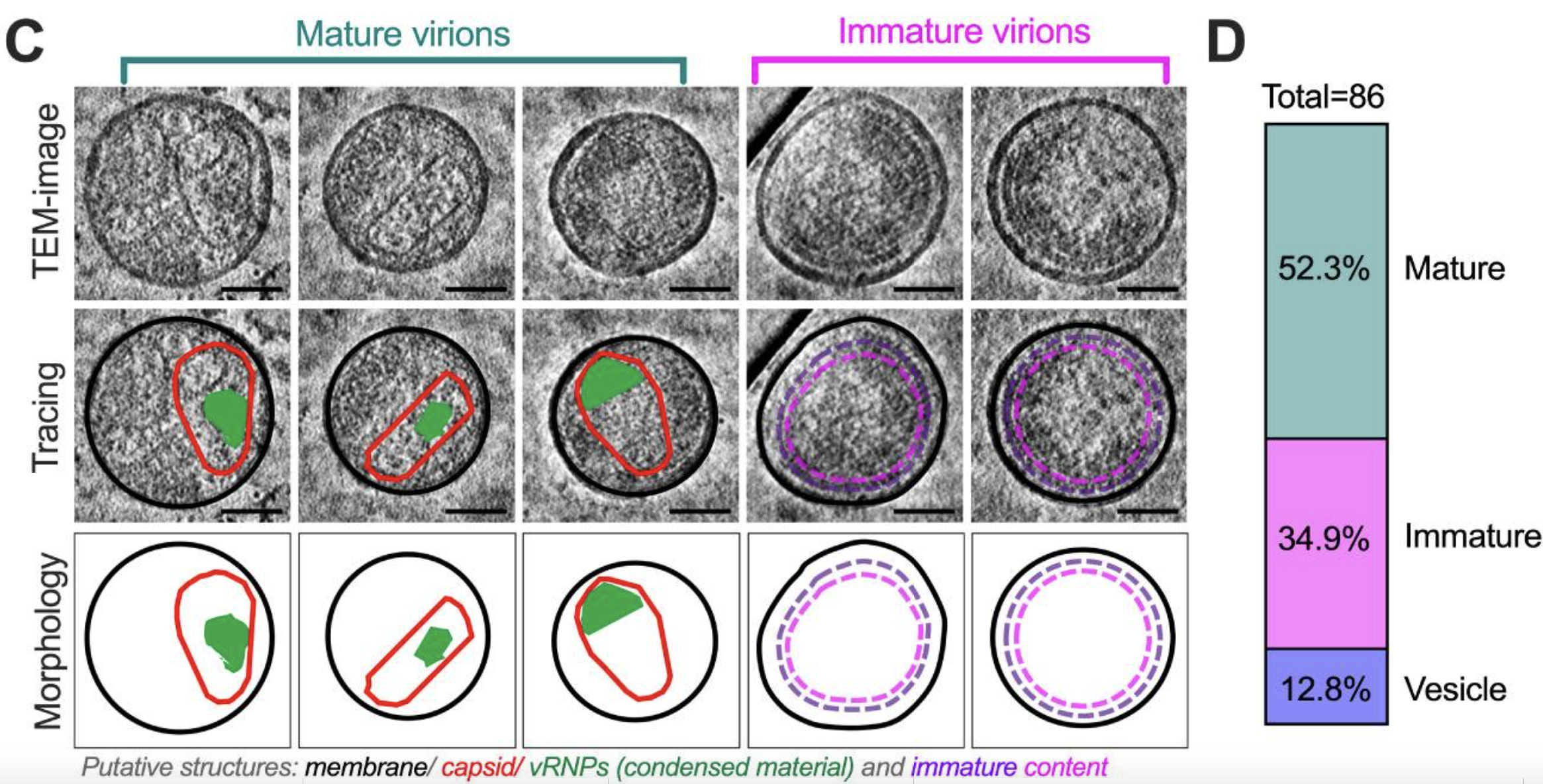
Dahmane S, Schexnaydre E, Zhang J, Rosendal E, Chotiwan N, Singh BK, Yau W, Lundmark R, Barad BA, Grotjahn DA, Liese S, Carlson A, Overby A, Carlson L. Cryo-electron tomography reveals coupled flavivirus replication, budding and maturation. bioRxiv 2024.
-
Access the paper
- PMID: 39416041
- Biorxiv Preprint: 618056
- Full Text Additional Links
- Carlson Lab
- Barad Lab
Klupt S, Fam KT, Zhang X, Chodisetti PK, Mehmood A, Boyd T, Grotjahn D, Park D, Hang HC. Secreted antigen A peptidoglycan hydrolase is essential for Enterococcus faecium cell separation and priming of immune checkpoint inhibitor cancer therapy. eLife 2024
CryoEM Methods
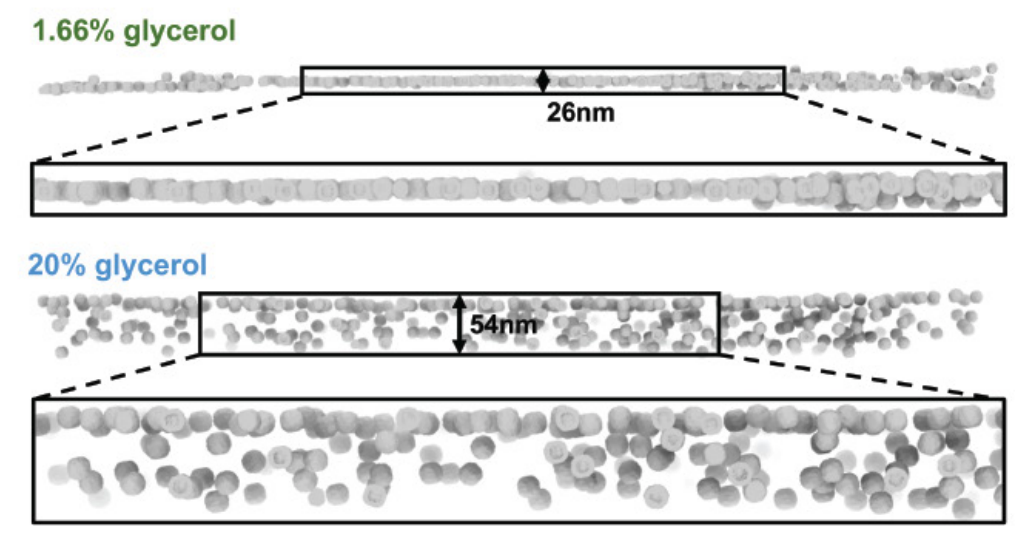
Basanta B, Hirschi MM, Grotjahn DA, Lander, GC. A case for glycerol as an acceptable additive for single particle cryoEM samples. Acta Crystallography D Structural Biology 2022
-
Access the paper
- PMID: 34981768
- PMCID: PMC8725161
- Biorxiv Preprint: 459874
- Full Text Additional Links
- Lander Lab
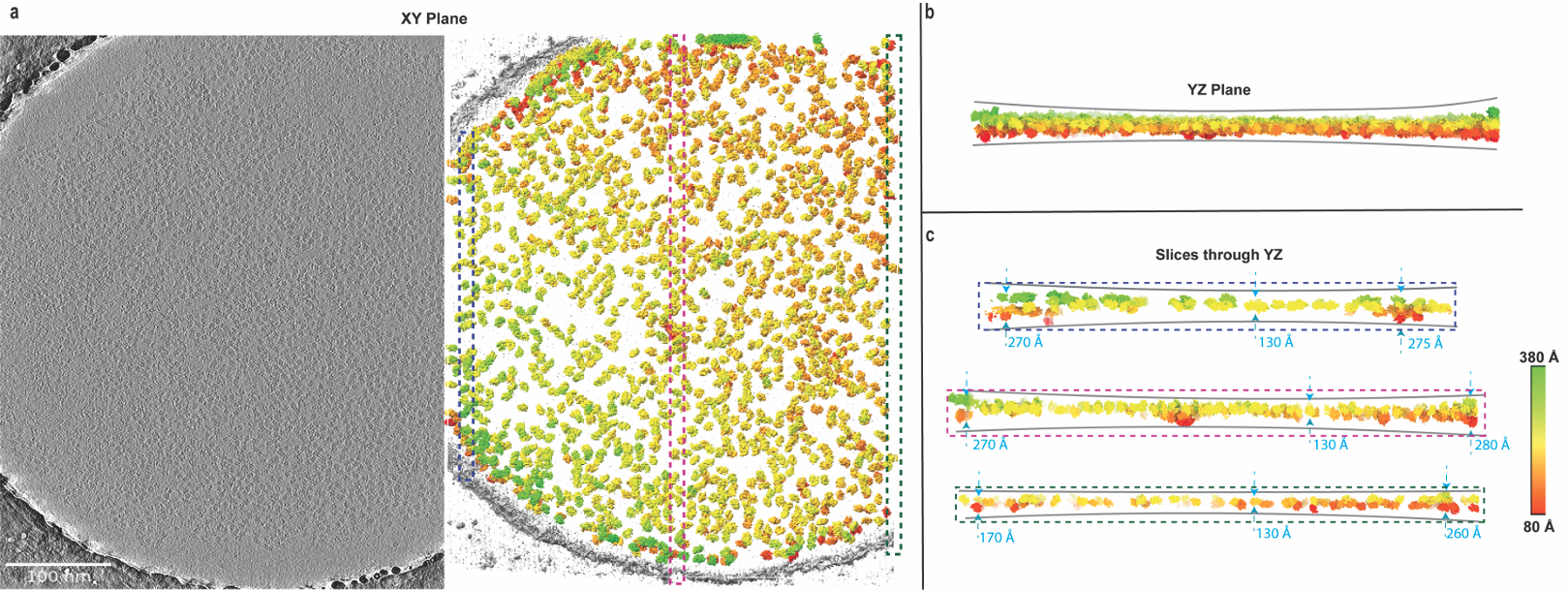
Aiyer A, Baldwin PR, Tan SM, Shan Z, Oh J, Mehrani A, Bowman ME, Louie ME, Passos DO, Đorđević-Marquardt S, Mietzsch M, Hull JA, Hoshika S, Barad BA, Grotjahn DA, McKenna R, Agbandje-McKenna M, Benner SA, Noel JAP, Wang D, Tan YZ, Lyumkis D.Overcoming resolution attenuation during tilted cryo-EM data collection. Nature Communications 2024
-
Access the paper
- PMID: 38195598
- Biorxiv Preprint: 548955
- Full Text Additional Links
- Lyumkis Lab
Collaborative Software Development
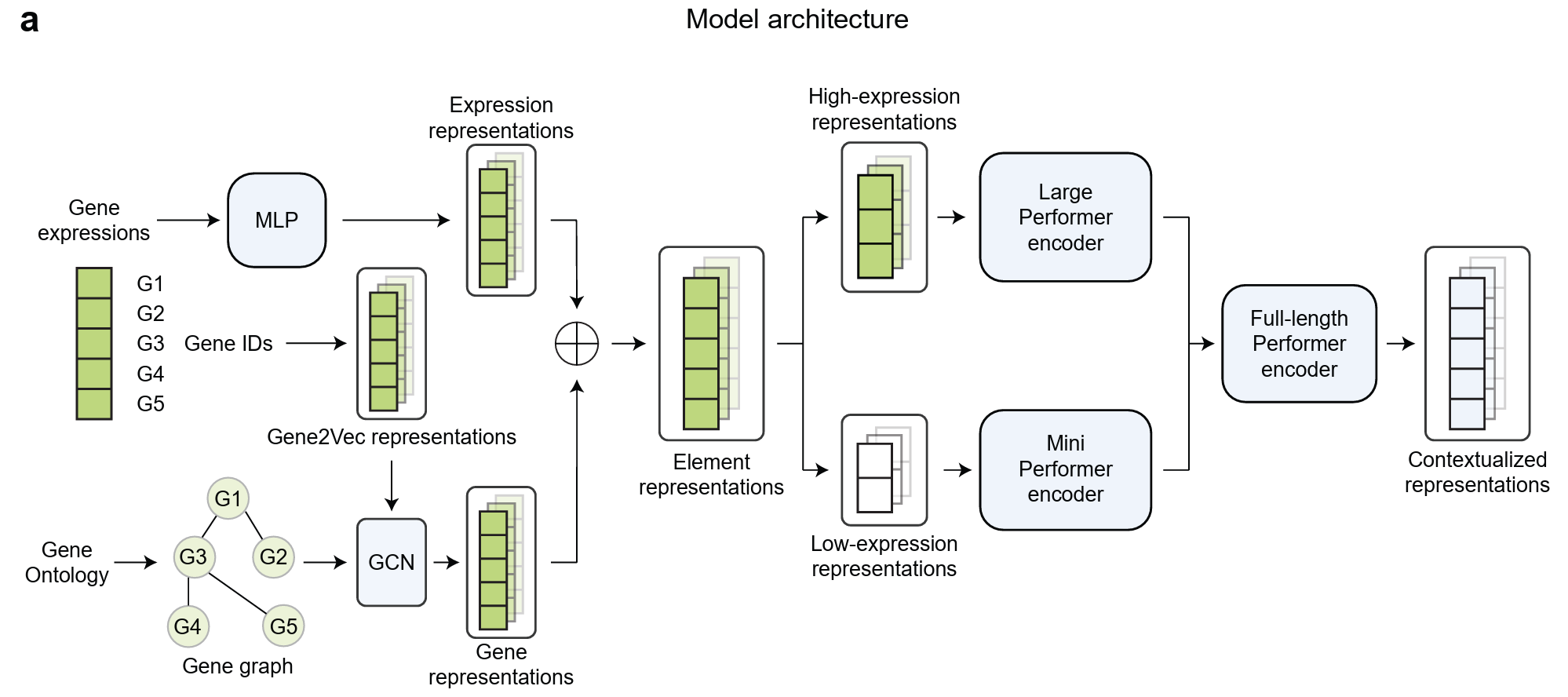
Bai D, Mo S, Zhang R, Luo Y, Gao J, Yang JP, Wu Q, Singh D, Rahmani H, Amariuta T, Grotjahn DA, Zhong S, Lewis N, Wang W, Ideker T, Xie P, Zing E. scLong: A Billion-Parameter Foundation Model for Capturing Long-Range Gene Context in Single-Cell Transcriptomics. bioRxiv 2024.
-
Access the paper
- PMID: XXXXX
- Biorxiv Preprint: 622759
- Full Text Additional Links
- Pengtao Xie Lab
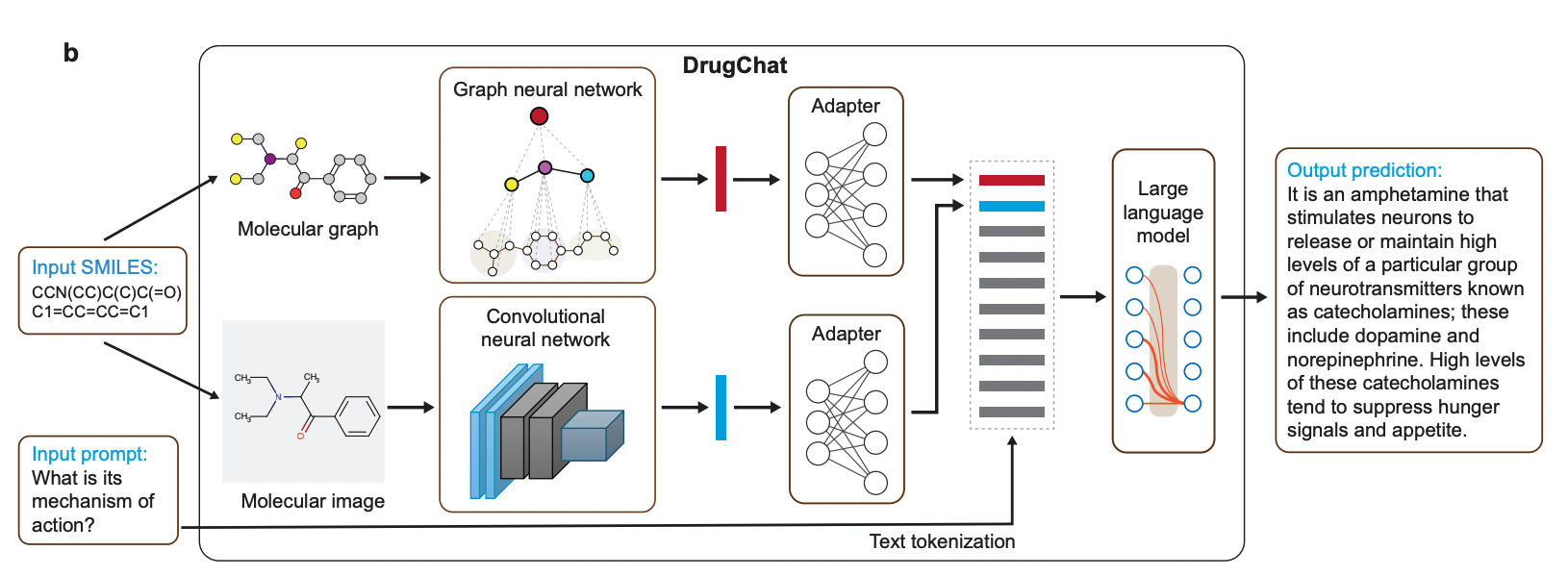
Liang Y, Zhang R, Li Y, Huo M, Ma Z, Singh D, Gao C, Rahmani H, Bandi S, Zhang L, Weinreb R, Malhotra A, Grotjahn DA, Awdishu L, Ideker T, Gilson M, Xie P. Multi-Modal Large Language Model Enables All-Purpose Prediction of Drug Mechanisms and Properties. bioRxiv 2024.
-
Access the paper
- PMID: XXXXX
- Biorxiv Preprint: 615524
- Full Text Additional Links
- Pengtao Xie Lab
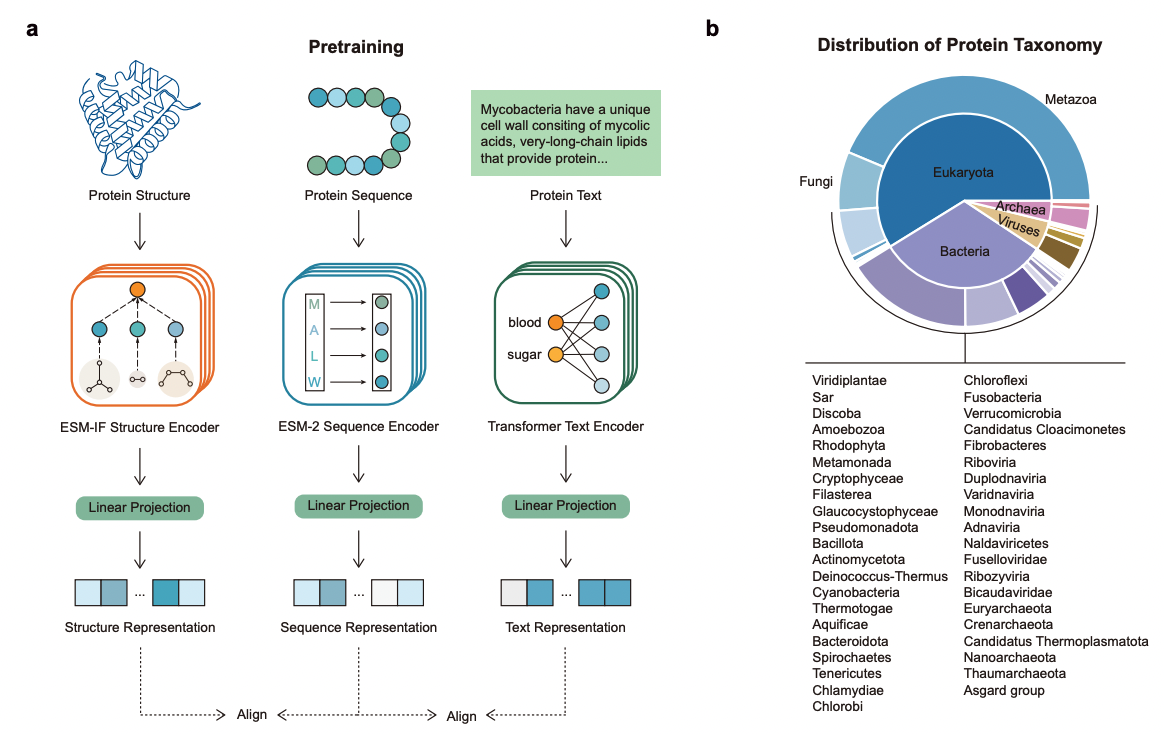
Zhang L, Guo H, Schaffer L, Ko YS, Singh D, Rahmani H, Grotjahn DA, Villa E, Gilson M, Wang W, Ideker T, Xing E, Xie P. ProteinAligner: A Multi-modal Pretraining Framework for Protein Foundation Models. bioRxiv 2024.
-
Access the paper
- PMID: XXXXX
- Biorxiv Preprint: 616870
- Full Text Additional Links
- Pengtao Xie Lab
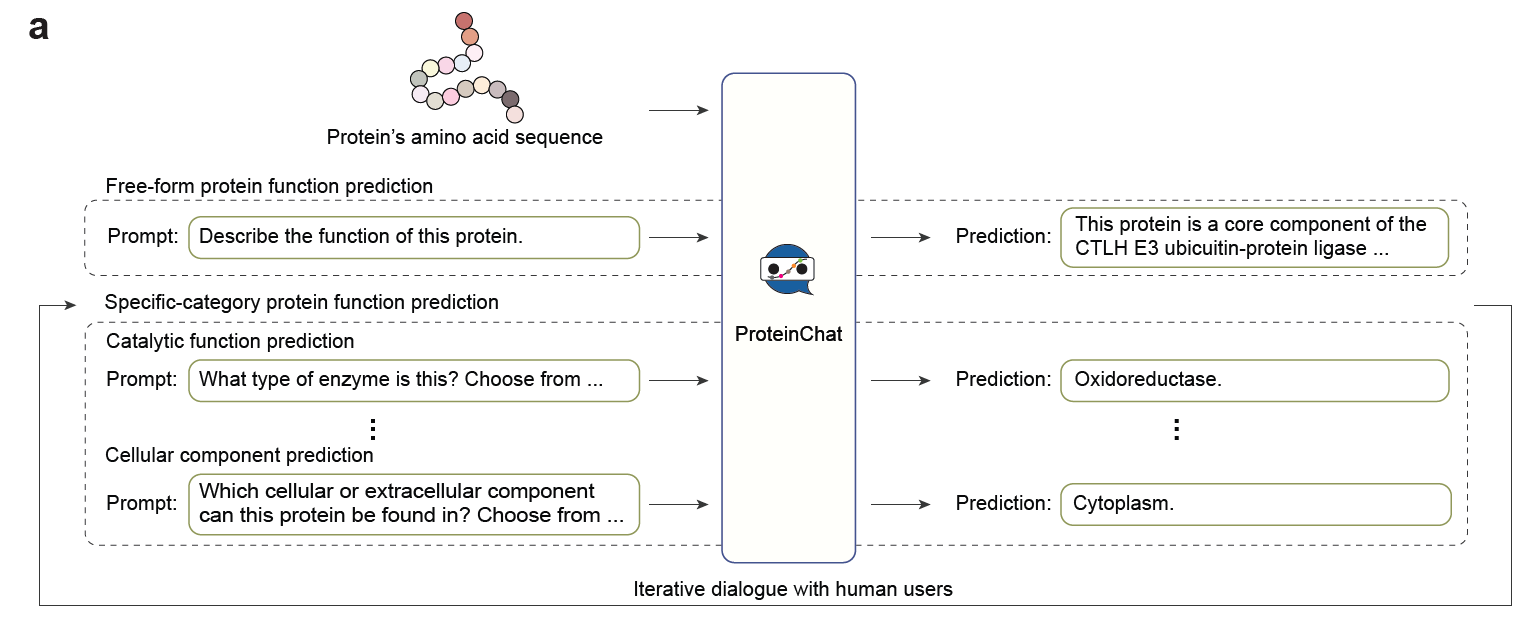
Huo M, Guo H, Cheng X, Singh D, Rahmani H, Li Shen, Gerlof P, Ideker T, Grotjahn DA, Villa E, Song L, Xie P. Multi-Modal Large Language Model Enables Protein Function Prediction. bioRxiv 2024.
-
Access the paper
- PMID: XXXXX
- Biorxiv Preprint: 608729
- Full Text Additional Links
- Pengtao Xie Lab